
RICCOMAGNO LAB
PROJECTS
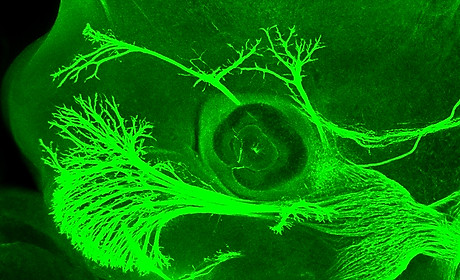
Mechanisms of signal integration
To form the right connections neurons must navigate long distances, guided by attractive and repulsive signals. These ‘guidance cues’ act like streetlights and street signs, and tell the developing neurons where to go to reach their final destinations. Just like when we are driving our cars, neurons encounter multiple signals along the road. While a number of these neuronal guidance signals have been identified, we still don’t understand how multiple attractive and repulsive cues are interpreted inside the developing neuron. Understanding how these cues are integrated and interpreted inside the cell to trigger a diverse array of developmental events is of critical importance if we are to understand the formation, function and malfunction of the human nervous system. We are investigating how a family of intracellular proteins participates in the establishment of brain circuits. These proteins (Cas family) have the potential to interpret and integrate the multiple signals that neurons receive.

Molecular mechanisms of pruning
The initial assembly of neuronal circuits is carried out by progressive developmental events, like the growth of axons (the main output projection of neurons). During these progressive or circuit-building events, excessive projections, which are unnecessary for the mature circuit, are formed. Subsequently, many brain circuits require refinement of these supernumerary connections via regressive events (e.g. death or devolvement of these byproduct projections) in order to properly function. One such regressive event, axonal pruning, remodels immature pathways by removal of exuberant axonal branches. Disruption of normal pruning events during neural development and circuit maturation has been linked to neurodevelopmental disorders, including autism-spectrum disorders. We are taking a variety of approaches to identify the molecular players critical for axonal pruning.

Targeting reactive astrocytes
Common to virtually all neurodegenerative diseases and brain disorders are changes in a glial cell type in the brain called an astrocyte, which become “reactive”. Astrocytes normally perform critical supportive functions in the brain, but the jury still out on whether reactive astrocytes are beneficial or detrimental for the progression of disease. In collaboration with the Fiacco and Wilson labs we are developing new approaches to study the role of reactive astrocytes during neurological disease.
LAB NEWS
-
We are looking for rotation students for the 2025-2026 academic year.
-
November 7th, 2025 - Our newest study is now published in PLOS Genetics.
-
April 9th, 2025 - Jason and Alyssa's genetic study is now a preprint in bioRxiv.
-
April 4th, 2025 - Congratulations to Payton on passing her Qualifying exam!
-
March 14th, 2025 - Congratulations to Alyssa on passing her Qualifying exam!
-
December 20th, 2024 - Teresa's paper describing detailed protocols for using the Lcn2CreERT2 mice is now published in STAR Protocols.
-
December 2024 - The Riccomagno Lab is awarded an R01 grant from NINDS. The main goal of this 5 year project is to determine the role of adhesion signaling and cytoplasmic adaptor proteins during cortical wiring.
-
July 10th, 2024 - Congratulations to Dr. Teresa on her successful thesis defense!
-
July 2024 - First NSF Surmount Summer Research Program is underway.
-
March 2024 - The Wilson, Fiacco and Riccomagno Labs are awarded a multi-PI R01 grant from NINDS to investigate the role of reactive astrocytes in toxoplasmosis.
-
September 7th, 2023 - Congratulations to Niloofar on her successful Master's Thesis defense!
-
August 4th, 2023 -Wenny and Jason's paper on an adhesion signaling pathway regulating the formation of the cortical glial scaffold is now published in Plos Biology.
-
July 1st, 2023 - Martin is now an Associate Professor.
-
May 25th, 2023 - Congratulations to Alyssa for being awarded a CIRM TRANSCEND Predoctoral Traineeship!
-
May 17th, 2023 - A revised version of Wenny and Jason's manuscript is now available as a preprint at bioRxiv.
-
April 13th, 2023 - The Riccomagno Lab is awarded a National Science Foundation Core grant. The main goal for this 5 year project is to understand the role of adhesion signaling during early cerebellar development.
-
April 4th, 2023 - Jason's paper is now published in Heliyon.
-
December 7th, 2022 - Congratulations to Jason on his successful thesis defense!
-
August 22nd, 2022 - Will and Teresa's paper on a new tool to target subsets of reactive astrocytes is now published in Cell Reports Methods. Here is a link. Now also featured here.
-
August 5th, 2022 - Wenny and Jason's work is now available as a preprint at bioRxiv.
-
December 8th, 2021 - Congratulations to Wenny on her successful thesis defense!
-
October 20, 2021 - Teresa and Tyler's new paper is published in the Journal of Cell Science, and got the cover! Check it out here.
-
July 28, 2021 - Congratulations to Will on his successful thesis defense!
-
December 15, 2020 - Congratulations to Tyler on his successful thesis defense!
-
July 30, 2020 - Our new preprint on the development of a viral-based approach for cell- and time-specific gene expression (ExBox) is now available at bioRxiv.
-
January 12, 2020 - Martin is now a Keystone Symposia Fellow.
-
We are looking for postdocs. See the ad here.
-
August 1, 2019 - The Fiacco, Riccomagno and Wilson Labs are awarded a multi-PI R01 grant from NIDA to investigate the role of reactive astrocytes in different models of chronic inflammation.
-
June 1, 2019 - The Riccomagno and Fiacco Labs are awarded an R03 grant from NIA to study the role of reactive astrocytes in Alzheimer's disease.
-
March 11, 2019 - Congratulations to Jason for passing his qualifying exam!
-
March 8, 2019 - The Riccomagno and Fiacco Labs are awarded an R21 grant from NINDS. The goal of the proposal is to develop a combinatorial strategy to selectively manipulate reactive astrocytes in disease.
-
January 25, 2019 - Congratulations to Wenny for passing her qualifying exam!
-
October 11, 2018 - The Riccomagno Lab is awarded a 1-year research grant from the CANCER RESEARCH COORDINATING COMMITTEE.
-
September 22, 2018 - The Riccomagno Lab is awarded an R21 grant from NIMH. The main goal of this project is to understand the role of caspase-dependent refinement during brain development.
-
August 29, 2018 - Congratulations to Tyler for passing his qualifying exam!
-
July 1st, 2018 - The Riccomagno Lab is awarded a Hellman Fellowship. The goal of the award is to explore the mechanisms that regulate thalamo-cortical axon pruning.
-
June 15, 2018 - The Riccomagno Lab is awarded an R01 grant from NINDS. The main goal of this 5 year project is to determine the role of adhesion signaling during cortical development.
-
Earlier 2018 - Congratulations to Jason and Tyler for publishing their papers in Scientific Reports!
PAST AND PRESENT FUNDING SOURCES







OUR TEAM
Because Science Is Fun!

Martin Riccomagno
Associate Professor

Alyssa Treptow
Graduate Student

Teresa Ubina
Graduate Student

Payton Depalma
Graduate Student

Tiffani Crenshaw
Graduate Student

Patrick Williamson
Lab Assistant

Sumukh Chanda
Underaduate Student
.jpg)
Yasamin Rahemi
Lab Assistant

Yiu-Cheung Wong
Lab Assistant Emeritus
FORMER TEAM MEMBERS
Graduate Students
Teresa Ubina - Postdoctoral fellow, UC Riverside
Jason Estep - Senior Scientist, Terremoto Biosciences
Wenny Wong - Scientist, Capricor Therapeutics, Inc.
Will Agnew-Svoboda - Senior Scientist, Arc Institute
Tyler Vahedi-Hunter - Lecturer at Cal State Bakersfield
Postdocs
Punit Bhattachan - Postdoctoral fellow at Albert Einstein College of Medicine
Lab assistants
Yiu-Cheung (Eric) Wong - PhD student at Stanford University
Camila Alvarez - PhD student at UC Riverside
Carly Horn - Clinical Laboratory Scientist Program
Undergraduate Students
Sunny Trieu - Pharmacy student of UC San Diego
Nahal Khalkhali (Honors) - Master's student at UC Irvine
Srinija Maganti - Postbac student at Dartmouth College
Natalie Taby (Honors) - Medical Assistant at Foothill Pediatric and Adolescent Clinic
Alexis Marquez - Lab Assistant at UC San Francisco
Kieusa Nguyen - Medical Student at Nova Southeastern University
Alexander Taft - Medical Student at Arkansas College of Osteopathic Medicine
Brian Loui - Medical student at Loma Linda University
Mandeep Chhokar - Postbac student at UC
Lauren Lopez (Honors) - Medical student at UC Riverside
Jasmine Pacheco - EMT
Jessica Avalos (MarcU)- Dental Student
Jonathan Argame -Master's student at Southern California University of Health Sciences
Anthony Chen - Clinical Genetic Molecular Biologist Scientist at Ambry Genetics
Abhinandan Singh Pabla - Medical Student at Stritch School of Medicine
THE LAB OVER THE YEARS






OUR TEAM
Because Science Is Fun!

Martin Riccomagno
Associate Professor

Alyssa Treptow
Graduate Student

Teresa Ubina
Graduate Student

Payton Depalma
Graduate Student

Camille Groneck
Graduate Student

Patrick Williamson
Lab Assistant

Sumukh Chanda
Undergraduate Student

Aria Olfati
Undergraduate Student

Sarah Parkinson
Undergraduate Student

Dania Khan
Undergraduate Student

Danny Haghighi
Undergraduate Student

Yiu-Cheung Wong
Lab Assistant Emeritus
FORMER TEAM MEMBERS
Graduate Students
Teresa Ubina - Postdoctoral fellow, UC Riverside
Jason Estep - Senior Scientist, Terremoto Biosciences
Wenny Wong - Scientist, Capricor Therapeutics, Inc.
Will Agnew-Svoboda - Senior Scientist, Arc Institute
Tyler Vahedi-Hunter - Lecturer at Cal State Bakersfield
Postdocs
Punit Bhattachan - Postdoctoral fellow at Albert Einstein College of Medicine
Lab assistants
Yiu-Cheung (Eric) Wong - PhD student at Stanford University
Camila Alvarez - PhD student at UC Riverside
Carly Horn - Clinical Laboratory Scientist Program
Undergraduate Students
Sunny Trieu - Pharmacy student of UC San Diego
Nahal Khalkhali (Honors) - Master's student at UC Irvine
Srinija Maganti - Postbac student at Dartmouth College
Natalie Taby (Honors) - Medical Assistant at Foothill Pediatric and Adolescent Clinic
Alexis Marquez - Lab Assistant at UC San Francisco
Kieusa Nguyen - Medical Student at Nova Southeastern University
Alexander Taft - Medical Student at Arkansas College of Osteopathic Medicine
Brian Loui - Medical student at Loma Linda University
Mandeep Chhokar - Postbac student at UC
Lauren Lopez (Honors) - Medical student at UC Riverside
Jasmine Pacheco - EMT
Jessica Avalos (MarcU)- Dental Student
Jonathan Argame -Master's student at Southern California University of Health Sciences
Anthony Chen - Clinical Genetic Molecular Biologist Scientist at Ambry Genetics
Abhinandan Singh Pabla - Medical Student at Stritch School of Medicine
THE LAB OVER THE YEARS






PUBLICATIONS
Estep, J.A.*, Treptow, A.M.*, Rao, P.A., Williamson, P., Wong, W., Riccomagno, M.M. (2025). Functional role for Cas cytoplasmic adaptor proteins during cortical axon pathfinding. PLoS Genet. 21(11):e1011941. PMC12611149
Ubina, T., Agnew-Svoboda, W., Figueroa, Z. A., Wilson, E. H., Fiacco, T. A., and Riccomagno, M. M. (2024) Protocol for the longitudinal study of neuroinflammation and reactive astrocytes in Lcn2CreERT2 mice. STAR Protocols 5(4): 103322. PMC11437937
Wong, W*, Estep, J.A*, Treptow, A.M. , Rajabli, N., Jahncke, J.N., Ubina, T., Wright, K.M., and Riccomagno, M.M. (2023) An adhesion signaling axis involving Dystroglycan, β1-Integrin and Cas adaptor proteins regulates the establishment of the cortical glial scaffold. Plos Biology 21(8): e3002212. PMC10431685
Estep, J.A, Sun, L.O., and Riccomagno, M.M. (2023) A Luciferase Fragment Complementation Assay to Detect Focal Adhesion Kinase (FAK) Signaling Events. Heliyon 9 (4): E15282. PMC10119766
Agnew-Svoboda, W., Ubina, T., Figueroa, Z., Wong, Y-C, Vizcarra, E., Roebini, B., Wilson, E., Fiacco, T.* and Riccomagno, M.M. * (2022) A genetic tool for the longitudinal study of a subset of post-inflammatory reactive astrocytes. Cell Reports Methods 2 (8) 100276. PMC9421582
Ubina, T.*, Vahedi-Hunter, T.*, Gupta, A., Wong, W., Agnew-Svoboda, W., Santhakumar, V., and Riccomagno, M.M. (2021) ExBoX: a simple Boolean exclusion strategy to drive expression in neurons. Journal of Cell Science 134 (20). PMC8572001
Rutlin, M., Rastelli, D., Kuo, W. T., Estep, J. A., Louis, A., Riccomagno, M. M., Turner, J. R., and Rao, M. (2020) The Villin1 Gene Promoter Drives Cre Recombinase Expression in Extraintestinal Tissues. Cellular and Molecular Gastroenterology and Hepatology 10:864-867. PMC7573669
Vahedi-Hunter, T.A., Estep, J.A., Rosette, K.A., Rutlin, M.L., Wright, K.M., and Riccomagno, M.M. (2018) Cas Adaptor Proteins Coordinate Sensory Axon Fasciculation. Scientific Reports 8:5996. PMC5902548
Estep, J.A., Wong, W., Wong, Y-C. E., Loui, B. M. and Riccomagno, M.M. (2018) β-Chimaerin regulates cerebellar granule cell development. Scientific Reports 8:680. PMC5766509
Agnew-Svoboda, W., Kolodkin, A.L.*, and Riccomagno, M.M.* (2016) Regressive Phenomena: Refining Connections. D.W. Pfaff, N.D. Volkow (eds.), Neuroscience in the 21st Century. * Authors for correspondence
Riccomagno, M.M. * and Kolodkin, A.L. *(2015) Sculpting Neural Circuits by Axon and Dendrite Pruning. Annual Review of Cell and Developmental Biology. 31: 779-805. * Authors for correspondence
Riccomagno, M.M.*, Sun, L. O.*, Brady, C.M., Alexandropoulos, K., Seo, S., Kurokawa, M., and Kolodkin, A.L. (2014) Cas adaptor proteins organize the Retinal Ganglion Cell Layer downstream of Integrin signaling. Neuron 81:779-786 * Equal contribution
Wang, S-H.J., Celic, I., Choi, S-Y., Riccomagno, M., Wang, Q., Sun, L.O., Mitchell, S., Vasioukhin, V., Huganir, R.L., and Kolodkin, A.L. (2014) Dlg5 Regulates Dendritic Spine Formation and Synaptogenesis by Controlling Subcellular N-cadherin Localization. Journal of Neuroscience 34: 12745-12761
Riccomagno, M.M., Hurtado, A., Wang H., Macopson, J.J, Griner, E.M., Betz, A., Brose, N., Kazanietz, M.G., and Kolodkin, A.L. (2012) The RacGAP β2-Chimaerin Selectively Mediates Axonal Pruning in the Hippocampus. Cell 149: 1594–1606
Pachikara, A., Dolson, D.K., Martinu, L., Riccomagno, M.M., Jeong, Y., and Epstein, D.J. (2007) Activation of Class I transcription factors by low level Sonic hedgehog signaling is mediated by Gli2-dependent and independent mechanisms. Developmental Biology 305: 52-62
Torban, E., Wang, H-J., Patenaude, A-M., Riccomagno, M., Daniels, E., Epstein, D., and Gros, P. (2006) Tissue, cellular and sub-cellular localization of the Vangl2 protein during embryonic development: effect of the Lp Mutation. Gene Exp. Patterns 7: 346-354
Riccomagno, M.M., Takada, S., and Epstein, D.J (2005) Wnt dependent regulation of inner ear morphogenesis is balanced by the opposing and supporting roles of Shh. Genes & Dev. 19: 1612-1623
Kleber, M., Lee, H-Y., Wurdak, H., Buchstaller, J., Riccomagno, M.M., Ittner, L.M., Suter, U., Epstein, D.J. and Sommer, L. (2005). Neural Crest Stem Cell Maintenance by Combinatorial Wnt and BMP Signaling. J. Cell. Bio. 169: 309-320
Riccomagno, M.M., Martinu, L., Mulheisen, M., Wu, D.K and Epstein, D.J. (2002) Specification of the mammalian cochlea is dependent on Sonic hedgehog. Genes & Dev. 16: 2365–2378
Paganelli, A., Ocaña, O., Prat, M. I., Franco, P., López, S., Morelli, L., Adamo, A., Riccomagno, M.M., Matsubara, E., Shoji, M., Affranchino, J., Castaño, E. and Carrasco, A. (2001) The Alzheimer-related gene presenilin-1 facilitates sonic hedgehog signalling in Xenopus primary neurogenesis. Mech. Dev. 107: 119-131
